Table of Contents
OrthoXML is an XML schema designed to describe orthology relations. Orthologs are defined as genes in different species deriving from a single gene in the last common ancestor. This relationship makes them interesting, as they are likely to have the same function.
OrthoXML is designed to be a versatile format to store orthology data from different sources in a uniform manner. It can store assignment from both pairwise approaches and tree based approaches with a variable level of detail. OrthoXML allows direct comparison and integration of orthology data from different resources. Additional, resource-specific information can also be included.
OrthoXML is a XML format. XML is a markup language which embeds the content in a structured way so that it easy to process and validate. Orthology data can be structured using XML as a container, where the relationships of genes and their orthology groups can be described as data objects. Since OrthoXML is defined as an XML schema, all XML files can be validated and checked to see if they are well-formed documents.
This document describes the content of OrthoXML version 0.2 and gives details about each element and attribute.
In this section we describe the structure of an OrthoXML file. We will not go through the whole schema piece by piece but rather show an example of a valid XML file.
The root element in the XML file is the orthoXML tag. It indicates also the version of the file, the origin of the data (database) and the version of the origin. Under the orthoXML tag one can find the tags notes, species, scores, and groups. The notes element contains additional information and is not validated. It can also be found under the elements species, groups, and geneRef.
A species element contains the basic information about the species, such as its name, the database where the gene records come from, and a list of those genes and their identifiers (IDs).
OrthoXML has no limitation regarding the number of species. Species pairwise orthology groups for multiple species can be stored in the same file. Furthermore, it supports multispecies ortholog groups, bifurcating trees or even multifurcating trees.
Different orthology databases use different types of scores for their assignments, and OrthoXML supports this. A typical score might be the confidence level for an ortholog group. Within the scores tag, multiple score types can be defined with scoreDef elements. Elsewhere in the file, orthology groups or group members have score values which correspond to the scoring schemes defined here. Since the score definitions are include in the OrthoXML files, scores from different resources can be uniformly processed.
The next element is the groups tag. This is the part of the file which contains the actual orthology assignments. The assignments are described as groups. There are two types of groups in OrthoXML: orthologGroups and paralogGroups. Both types can contain both genes and nested groups. The genes that are members of a group are denoted using geneRef tags that reference the genes defined earlier in the species section. Members of an OrthologGroup are meant to be orthologous to each other, whereas members of a paralogGroup are paralogous to each other. On the top level of the groups tag, only OrthologGroups are allowed since the purpose of OrthoXML is to to store orthology relations. By nesting group elements, phylogenetic relationships can be repesented. More details and examples how to store different trees are shown in the next section, "Trees". Property tags nested in a group tag allow key-value annotations .
<?xml version="1.0" encoding="utf-8"?>
<orthoXML xmlns="http://orthoXML.org/2011/" version="0.3" origin="inparanoid"
originVersion="7.0" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:schemaLocation="http://orthoXML.org/2011/ http://www.orthoxml.org/0.3/orthoxml.xsd">
<notes>
Example OrthoXML file. Stripped down version of a real InParanoid 7.0 file.
</notes>
<species name="Caenorhabditis elegans" NCBITaxId="6239">
<database name="WormBase" version="Caenorhabditis-elegans_WormBase_WS199_protein-all.fa"
geneLink="http://www.wormbase.org/db/gene/gene?name="
protLink="http://www.wormbase.org/db/seq/protein?name=WP:">
<genes>
<gene id="1" geneId="WBGene00000962" protId="CE23997" />
<gene id="5" geneId="WBGene00006801" protId="CE43332" />
</genes>
</database>
</species>
<species name="Homo Sapiens" NCBITaxId="9606">
<database name="Ensembl" version="Homo_sapiens.NCBI36.52.pep.all.fa"
geneLink="http://Dec2008.archive.ensembl.org/Homo_sapiens/geneview?gene="
protLink="http://Dec2008.archive.ensembl.org/Homo_sapiens/protview?peptide=">
<genes>
<gene id="2" geneId="ENSG00000197102" protId="ENSP00000348965" />
<gene id="6" geneId="ENSG00000198626" protId="ENSP00000355533" />
<gene id="7" protId="ENSP00000373884" />
</genes>
</database>
</species>
<scores>
<scoreDef id="bit" desc="BLAST score in bits of seed orthologs" />
<scoreDef id="inparalog" desc="Distance between edge seed ortholog" />
<scoreDef id="bootstrap" desc="Reliability of seed orthologs" />
</scores>
<groups>
<orthologGroup id="1">
<score id="bit" value="5093" />
<property name="foo" value="bar"/>
<geneRef id="1">
<score id="inparalog" value="1" />
<score id="bootstrap" value="1.00" />
</geneRef>
<geneRef id="2">
<score id="inparalog" value="1" />
<score id="bootstrap" value="1.00" />
</geneRef>
</orthologGroup>
<orthologGroup id="3">
<score id="bit" value="3795" />
<geneRef id="5">
<score id="inparalog" value="1" />
<score id="bootstrap" value="1.00" />
</geneRef>
<geneRef id="6">
<score id="inparalog" value="1" />
<score id="bootstrap" value="1.00" />
</geneRef>
<geneRef id="7">
<score id="inparalog" value="0.4781" />
</geneRef>
</orthologGroup>
</groups>
</orthoXML>
OrthoXML can as stated above represent both flat and hierarchical ortholog groups. The example given above shows two simple ortholog groups under the groups tag. To represent a hierarchical (that is, phylogenetic) relationship between those groups, one can nest paralogGroups or othologGroups under the top level orthologGroup tag. Notice that the highest level group tag must always be an orthologGroup, and each group must contain at least two nested elemets of the type orthologGroup, paralogGroup or geneRef. GeneRef are the members of the of the group (or leaves if you think in terms of trees). The top level ortholog group is the root of the tree and nested groups are child nodes of their parent group. Currently, OrthoXML supports the two most common events in phylogenies — duplications, which are represented as paralogGroups, and speciations, in the form of orthologGroups. The level of detail in which the trees are given is up to the user. For example, nodes can be omitted and sets of leaves can be placed in one group. Below are some simple examples which demonstrate how different phylogenies can be represented in OrthoXML.
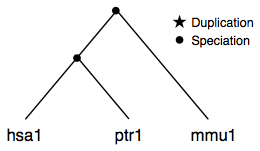
A simple case with three genes from different species that are descendants of a single gene in their last common ancestor. This is represented as an orthologGroup containing another nested orthologGroup and a single geneRef.
<?xml version="1.0" encoding="utf-8"?>
<orthoXML xmlns="http://orthoXML.org/2011/" version="0.3" origin="orthoXML.org" originVersion="1">
<species name="Homo sapiens" NCBITaxId="9606">
<database name="someDB" version="42">
<genes>
<gene id="1" geneId="hsa1" />
</genes>
</database>
</species>
<species name="Pan troglodytes" NCBITaxId="9598">
<database name="someDB" version="42">
<genes>
<gene id="2" geneId="ptr1"/>
</genes>
</database>
</species>
<species name="Mus musculus" NCBITaxId="10090">
<database name="someDB" version="42">
<genes>
<gene id="3" geneId="mmu1"/>
</genes>
</database>
</species>
<groups>
<orthologGroup>
<orthologGroup>
<geneRef id="1"/>
<geneRef id="2"/>
</orthologGroup>
<geneRef id="3"/>
</orthologGroup>
</groups>
</orthoXML>
|
|
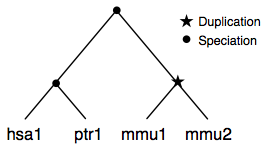
A slightly more complex example where a duplication has occured in one species. This results in the introduction of a paralogGroup.
<?xml version="1.0" encoding="utf-8"?>
<orthoXML xmlns="http://orthoXML.org/2011/" version="0.3" origin="orthoXML.org" originVersion="1">
<species name="Homo sapiens" NCBITaxId="9606">
<database name="someDB" version="42">
<genes>
<gene id="1" geneId="hsa1"/>
</genes>
</database>
</species>
<species name="Pan troglodytes" NCBITaxId="9598">
<database name="someDB" version="42">
<genes>
<gene id="2" geneId="ptr1"/>
</genes>
</database>
</species>
<species name="Mus musculus" NCBITaxId="10090">
<database name="someDB" version="42">
<genes>
<gene id="3" geneId="mmu1"/>
<gene id="4" geneId="mmu2"/>
</genes>
</database>
</species>
<groups>
<orthologGroup>
<orthologGroup>
<geneRef id="1" />
<geneRef id="2" />
</orthologGroup>
<paralogGroup>
<geneRef id="3" />
<geneRef id="4" />
</paralogGroup>
</orthologGroup>
</groups>
</orthoXML>
|
|
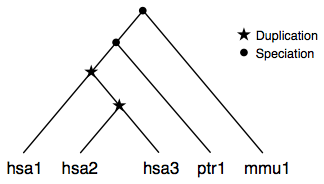
In this example, multiple duplication have occurred in the human lineage after the differentiation from chimpanzee. We don't want to specify the relationship of the human genes to each other, so we have placed them into a single paralogGroup.
<?xml version="1.0" encoding="utf-8"?>
<orthoXML xmlns="http://orthoXML.org/2011/" version="0.3" origin="orthoXML.org" originVersion="1">
<species name="Homo sapiens" NCBITaxId="9606">
<database name="someDB" version="42">
<genes>
<gene id="1" geneId="hsa1"/>
<gene id="2" geneId="hsa2"/>
<gene id="3" geneId="hsa3"/>
</genes>
</database>
</species>
<species name="Pan troglodytes" NCBITaxId="9598">
<database name="someDB" version="42">
<genes>
<gene id="4" geneId="ptr1"/>
</genes>
</database>
</species>
<species name="Mus musculus" NCBITaxId="10090">
<database name="someDB" version="42">
<genes>
<gene id="5" geneId="mmu1"/>
</genes>
</database>
</species>
<groups>
<orthologGroup>
<orthologGroup>
<paralogGroup>
<geneRef id="1" />
<geneRef id="2" />
<geneRef id="3" />
</paralogGroup>
<geneRef id="4" />
</orthologGroup>
<geneRef id="5" />
</orthologGroup>
</groups>
</orthoXML>
|
|
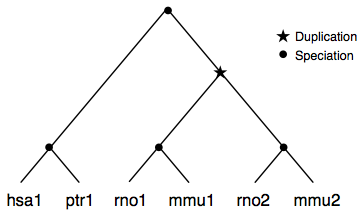
More complex example where a duplication has occured before the separation of mouse and rat resulting in a paralogGroup with two nested orthologGroups.
<?xml version="1.0" encoding="utf-8"?>
<orthoXML xmlns="http://orthoXML.org/2011/" version="0.3" origin="orthoXML.org" originVersion="1">
<species name="Homo sapiens" NCBITaxId="9606">
<database name="someDB" version="42">
<genes>
<gene id="1" geneId="hsa1" protId="hsa1" />
</genes>
</database>
</species>
<species name="Pan troglodytes" NCBITaxId="9598">
<database name="someDB" version="42">
<genes>
<gene id="2" geneId="ptr1"/>
</genes>
</database>
</species>
<species name="Mus musculus" NCBITaxId="10090">
<database name="someDB" version="42">
<genes>
<gene id="3" geneId="mmu1"/>
<gene id="4" geneId="mmu2"/>
</genes>
</database>
</species>
<species name="Rattus norvegicus" NCBITaxId="10116">
<database name="someDB" version="42">
<genes>
<gene id="5" geneId="rno1"/>
<gene id="6" geneId="rno2"/>
</genes>
</database>
</species>
<groups>
<orthologGroup>
<orthologGroup>
<geneRef id="1" />
<geneRef id="2" />
</orthologGroup>
<paralogGroup>
<orthologGroup>
<geneRef id="3" />
<geneRef id="5" />
</orthologGroup>
<orthologGroup>
<geneRef id="4" />
<geneRef id="6" />
</orthologGroup>
</paralogGroup>
</orthologGroup>
</groups>
</orthoXML>
|
|
February 23, 2011
-
Namespace change: http://orthoXML.org/0.2 → http://orthoXML.org/2011/
This makes the namespace independent from version changes. From now on namespace changes will be reserved to major schema changes.
A property tag was added to the ortholog and paralog group tags to support key-value annotations.
The gene identifier of the gene element is now optional similar to the protein identifier and transcipt identifier. Please note that a meaningful gene element should have at least one of the three.
November 2, 2010
-
Renaming of the cluster tags to group tags.
clusters → groups
cluster → orthologGroup
New element paralogGroup.
Nesting of groups to represent phylogentic trees.
The id attribute from species was removed.
The type of orthoXML/@originVersion changed to xs:token.
-
Requirement constraint for some attributes and elements were removed.
orthoXML/scores
orthologGroup/@id
orthologGroup/score
geneRef/score